Informator uniwersytecki
OPUS 21 - dr Matthias Nees
W ramach konkursu OPUS 21 dofinansowanie otrzymał projekt pt.: „Analiza porównawcza systemów hodowli tkanek ludzkich, które naśladują mikrośrodowisko guza”. Kierownikiem projektu jest dr Matthias Nees, prof. uczelni z Katedry i Zakładu Biochemii i Biologii Molekularnej. Projekt realizowany jest we współpracy z Uniwersytetem Warszawskim oraz Instytutem Chemii Fizycznej PAN.
Looking Deep into the Core of Cancer Tissues
Cancer is a highly personal disease – each patient’s tumour is a unique and complex living tissue, that reacts differently to therapies. For aggressive cancer types, such as head & neck (H&N) cancer, there is no cure for up to 50% of the patients. Response to therapies only partly depends on the malignant cancer cells themselves. Recent research, however, points to the very critical role of the “tumour microenvironment” (TME). New concepts consider the TME as a fundamental and functional component of any tumour – and it is individually very different from patient to patient. One of the most important components of the TME are the stromal cells, or fibroblasts. These cells, common to almost every healthy tissue, can be turned by cancer cells into cancer-associated fibroblasts or CAFs. These CAFs now strongly support and protect the tumour cells in all stress situations. Another important component are immune cells which often invade into the tumour tissue. Immune cells like T-cells and Natural Killer or NK cells can actively attack and fight tumour cells. However, in many patients, the tumour has developed mechanisms that allows the cells to escape this cellular immune response, which leads to immune resistance. Stromal cells or CAFs are one driving force behind therapy failure, and immune resistance. It even seems that stromal fibroblasts may actively prevent the invasion of immune cells into tumour tissues. This generates an immunosuppressive tumour microenvironment, sometimes called the tumour immune microenvironment, or TIME. Another important aspect is blood circulation within tumours, and the formation of capillaries, or new blood vessels in the tumour tissue. This is accomplished by endothelial cells. These cells also communicate with tumour, stromal and immune cells, and may contribute to shaping an immunosuppressive TME. In this project, we aim to investigate how this communication between different cells may be responsible for the failure or success of therapies.
Immuno-Oncology and Personalized Medicine: One important aspect of our research is the immune system. Tumours that can effectively avoid the invasion of immune cells are often called “cold” tumours. In contrast, “hot” tumours may contain different types of immune cells, such as T- and B-cells, or natural killer (NK) cells. As a rule, only “hot” tumours respond to immunotherapies. They also show a generally better response to any other form of therapy used in the clinic, including irradiation and different forms of chemotherapy (such as cisplatin and Taxanes). How exactly the tumour may or may not silence the immune response is still poorly understood, and it is extremely difficult to predict if a tumour will respond or not to a certain therapy. This is now one of the most important questions for personalized cancer medicine (PCM). Personalized medicine aims for better prediction of therapy outcome in patients. It also aims at a better classification of patients into groups that would be most likely respond to certain therapies (also known as “patient stratification”). PCM, however, remains difficult and there are almost no tools to predict if head & neck cancer patients may respond to which therapies.
Immune Checkpoint Inhibitors: New forms of therapies have been developed during the last decade, which target the immune system. These therapies can re-activate the invasion of immune cells into tumour tissues, where they start to attack the tumour cells. Immunotherapies may also change an immunosuppressive “cold” TME into a more immuno-reactive “hot” TME. The most successful immunotherapies are the immune checkpoint inhibitors (ICI), which help to save the lives of many patients. However, only a fraction of patients (around 20-30%) shows a response. A better understanding of the communication between tumour and other cells would help to better predict how these immunotherapies work – of why they fail in so many patients. The complex composition of the TME and the different cell types constantly “talking” to each other may be one of the most relevant factors for predicting if certain immunotherapies may work, and for which patients. Understanding this language would be highly beneficial for personalized medicine and could help to avoid inefficient and costly therapies – with unnecessary suffering of the patients.
Metastasis: Tumour and non-malignant cells in the TME also form relationships, which are critical for metastasis – the spread of cancer to distant parts of the body. Metastases in head & neck cancers are usually not incurable, and eventually lead to the death of the patients. The TME may also be relevant for the observation that cancers often “come back” or relapse, many years after therapies. This phenomenon is called “tumour dormancy”: tumour cells can be completely inactive or “sleep”, hidden in the patients tissues - but then suddenly become active again. This can happen years or decades after therapy. Such recurrent tumours are also often incurable, and tend to be much more aggressive than the initial cancer.
Model systems for cancer research are all incomplete: The role of the TME as an essential component of tumours has been underestimated in cancer research since the beginnings. The TME has even been considered by many as completely irrelevant for cancer progression or the failure of therapies. Although this is now changing, the most important problem is that there are only very few model systems for cancer research that contain anything else but the tumour cells. At the same time, research largely depends on the availability of models, and their quality. For example, model systems without immune cells are useless for any meaningful research on immune-oncology, and immune checkpoint inhibitors. Similarly, models without vasculature cannot possibly be useful to investigate the formation of new blood vessels in cancers, a process called angiogenesis. Most of the in vitro (or ex vivo) models that are currently used in cancer research, even the more advanced ones like tumour “organoids”, do not include any of the non-malignant cell types found in the TME. But the same is true even for animal models: the transplantation of tumour cells or small pieces of tumour tissue into immunocompromised mice (a technique called xenografting) has been a common strategy to study human tumours. Yet, tumours that are formed in this way are still derived exclusively from monoclonal cancer cells. These xenografts do not reflect the true heterogeneity of real tumours, and typically do not include any immune cells.
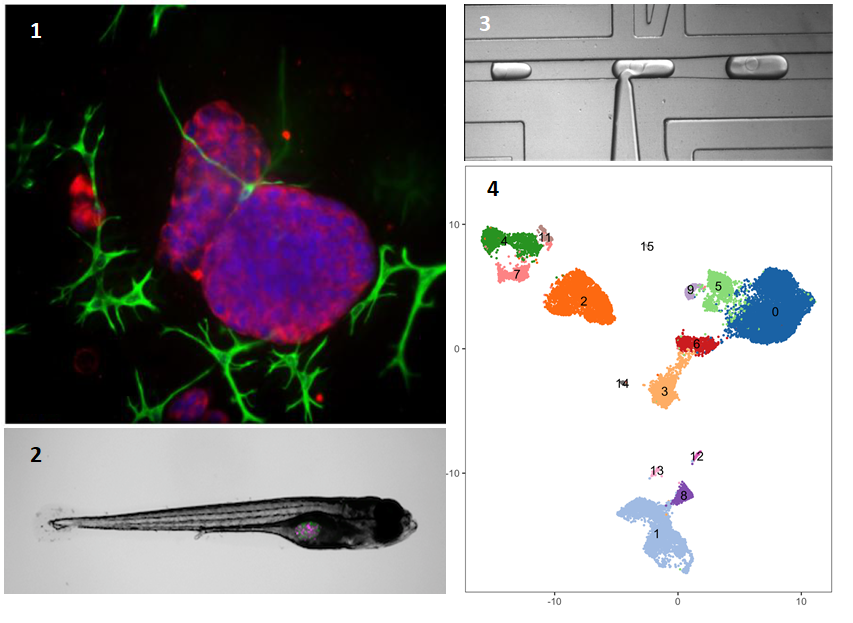
Improved in vitro model systems for cancer biology: This new project, funded by NCN, aims to develop improved model systems, which are not formed only by the human cancer cells. We will use advanced cell- and tissue culture strategies, or tissue engineering, to establish better cancer models.At the same time, we will not use tumour cell lines – often established decades ago. Instead, we will use fresh tumours from patients, to extract living tumour and supporting cells, including immune cells. This research will be conducted at the Medical University of Lublin (UMLUB), Department of Biochemistry & Molecular Biology, and is coordinated by Matthias Nees. Our tumour models will be as complex as possible – and will contain all the critical components of patient tissues. We will use two opposing strategies for this goal: 1) we will isolate all the cells from a patients’ tumour tissue as a “cell suspension”, and culture them in different experimental systems. That a) will allow them to again form tissue-like structures also known as organoids; and b) help us to maintain the heterogeneity of a tumour tissue. Such models are now known as “patient-derived organoids” (PDOs). Our advanced version will also contain cells from the immune system and the vasculature. For this goal, we will utilize methods originally developed for tissue engineering. Figure panel 1 shows such a tumour organoid, cultured in extracellular matrix – the scaffold surrounding all cells within a tissue. Here, the organoid (red) is surrounded by cancer-associated fibroblasts (green). 2) In another, alternative model system, we will isolate small pieces or slices of tissue from the original tumour biopsy, and culture them on scaffolds that support their growth for a prolonged period (at least 1-2 weeks). These “patient-derived explants”, or PDEs, naturally contain all of the different cells found in the original tissue; including the immune cells and the stromal fibroblasts. 3) In addition, we will explore how the heterogeneity of patient-derived cells and tissues (as above) is maintained when transplanted (or xenografted) into zebrafish (Figure panel 2). The research using zebrafish is also performed at UMLUB, coordinated by Adolfo Rivero-Müller.
Single-cell RNA Sequencing: All three model systems will be developed in parallel. This will make it possible for us to address how the tumour tissue forms and how it responds to various therapies. We will use the emerging technology of single-cell RNA sequencing to observe which cells are contained in these tumour microtissues, and how the composition may change during and after therapies. This is a novel technique in which single cells are first isolated from different tissues (or tissue models, in our case), and analysed by DNA sequencing in the next step. Isolation of single cells will be achieved by the unique and customizable droplet microfluidic platform developed by Tomasz Kaminski at the University of Warsaw (UW). This highly effective method allows us to separate and encapsulate millions of single cells into tiny droplets (Figure panel 3). In the next step, the single cells, each captured in a nanolitre droplet are lysed. Messenger RNA (mRNA) is then released from cytoplasm and nuclei, and reverse-transcribed into copy-DNA (cDNA). Each of the tiny droplets carries a oligonucleotide "barcode" that uniquely labels the cDNA derived from each cell. This barcoding allows us to identify from which cell the mRNA/cDNA originally came. Once reverse transcription is completed, the droplets can be fused into one large reaction volume for further preparation of so-called DNA libraries. These are finally sequenced by next-generation DNA sequencing (NGS) methods. In this fashion, we can sequence millions of cells originally isolated from either tumour tissues, or the models and identify the nature of each individual cell (Figure panel 4).
Bioinformatics: Characteristic gene signatures are then used to identify tumour and stromal cells, and to distinguish them, for example, from the different immune cells that are likely present in both models and the original cancer tissues. For this, we will use advanced bioinformatics methods, developed originally at MIT in Boston/USA. In Figure panel 4, the “clusters” in different colours each represent such a different cell type, originally isolated from a single cancer tissue. Bioinformatics will allow us to not only identify, but also quantify the numbers of such cell populations. The bioinformatics part will be coordinated by Marcin Tabaka (ICTER/Institute of Physical Chemistry of Polish Academy of Sciences, Warsaw). The goal is to identify any changes in the cellular composition of tumour tissues before, during and after therapies. Understanding the composition of the models will then also allow us to test if the same processes, cell types, and dynamic changes may also be critical for the response of patient’s tumours to therapies.
Functional validation of our findings and clinical relevance: By looking deeply into the cell composition of tumours and measuring changes in gene expression of different cell types found in these tumours, we can test if any of these changes are associated with the response or failure of therapies in patients. This is a unique opportunity to test scientific hypotheses and may help us understanding therapy failures in patients. It may also be useful to identify genes, or certain patterns of gene expression (so-called “gene signatures”) or biomarkers, that predict therapy responses. Identifying patients that are likely to respond to certain therapies, but not to others, may result in improving the the efficacy of therapies, and avoid unnecessary treatments that are not likely to be beneficial. This is an important step to improve the quality of life (QoL) of patients, which are often suffering from the side effects of certain therapies, such as radiation or cisplatin. The long-term goal of our research is to improve future personalized medicine, and help clinicians make informed decisions concerning how a particular patient should be treated (individualized therapy).
Author: Matthias Nees






